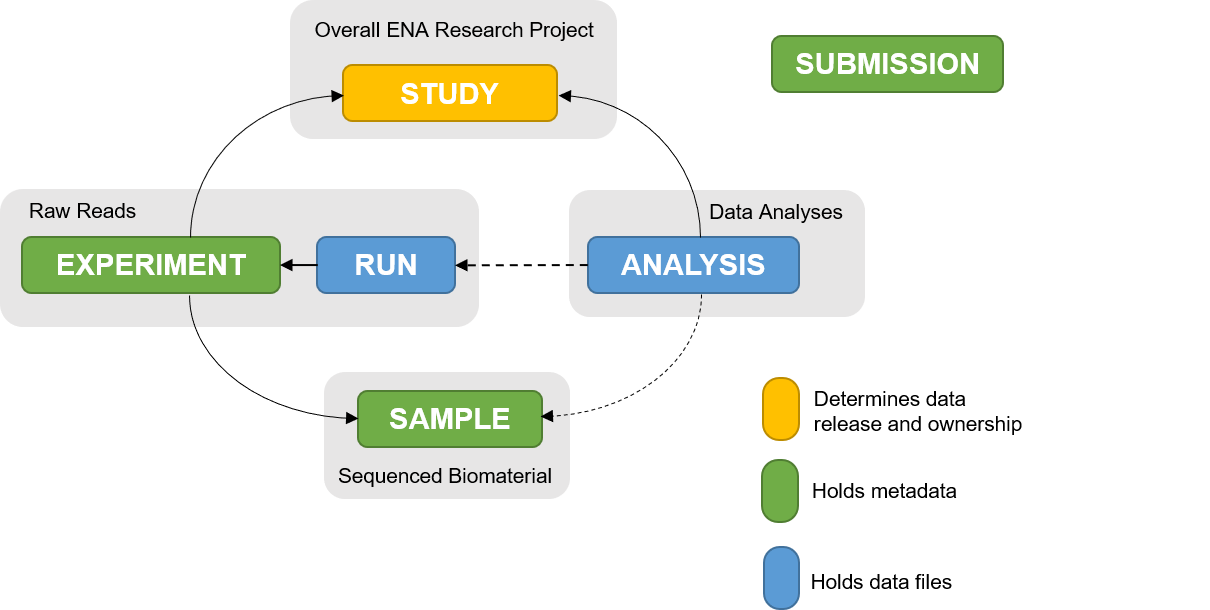
The ENA Metadata model foresees the following elements to be uploaded:
To support the uploading process the ENA Upload Microservice (ena_upload_ms) is built. You can register upload jobs of data and metadata and the microservice will handle the jobs in the background and updates the job information with additional data that is received by the ena upload procedure.
Mainly there are three different uploads available:
- Study definition upload (
/jobs/study) - Sample-Experiment-Run upload (
/jobs/ser) - Analysis upload (
/analysisjobs)
Often the majority of metadata for one project are the same for all upload jobs. Therefore you can add a project related template that contains all the metadata that is static during the project. This information is stored in a yaml file in the templates directory:
checklist: ERC000033
center_name: My Center
laboratory: My Laboratory
# Overall ENA research project
study:
alias: my_study_alias
title: My Study Title
study_type: studyType
study_abstract: My Study Abstract
pubmed_id: pubmedId (if available)
# Sequenced Biomaterial
sample:
alias: my_sample_{}
title: My Sample Title
taxon_id: The Taxon ID
sample_description: My Sample Description
host subject id: 0000001
collection date: 2023-11-27
geographic location (country and/or sea): Switzerland
host common name: Human
host health state: "missing: human-identifiable"
host sex: "missing: human-identifiable"
host scientific name: Homo Sapiens
collector name: ""
collecting institution: Collecting institution
isolate: ""
# Raw Reads
experiment:
alias: my_experiment_{}
title: My Experiment
study_alias: my_study_alias
sample_alias: my_sample_{}
design_description: "None"
library_name: "None"
library_strategy: RNA-Seq
library_source: METAGENOMIC
library_selection: cDNA
library_layout: "paired"
insert_size: 250
library_construction_protocol: "None"
platform: illumina
instrument_model: Illumina MiSeq
# Files
run:
alias: my_run_{}
experiment_alias: my_experiment_{}
# Data Analysis
analysis:
name: my_analysis_{}
assembly_type: clone
coverage: <float with the coverage>
program: TODO
platform: TODOThe {} will be replace with a timestamp of nanoseconds accuracy (strftime("%Y%m%d%H%M%S%f")). Before you can create an analysis job you need to create a job, preferably including a Sample-Experiment-Run component. The Analysis Job will then point (Foreign key) to this job. This is because you need to have reference a run and a sample from the analysis and thats only possible if they already exist. That leads to the following order of uploads:
- Upload the study (this is needed only once):
/jobs/study/ - Upload Sample-Experiment-Run-Tripple:
/jobs/ser/ - Upload an Analysis:
/analysisjobs/ - Attach files to the analysis job
/analysisfiles/ - Enqueue the analysis job
- Release the job
/jobs/<job_id>/release - Release the analysisjob
/analysisjobs/<job_id>/release
version: "3"
volumes:
pg_data:
services:
ena:
image: ethnexus/ena-upload-ms
hostname: ena
restart: unless-stopped
volumes:
- ./ena/templates:/templates
- ./data:/data
environment:
- POSTGRES_HOST=${POSTGRES_HOST}
- POSTGRES_PORT=${POSTGRES_PORT}
- POSTGRES_DB=${ENA_POSTGRES_DB}
- POSTGRES_USER=${ENA_POSTGRES_USER}
- POSTGRES_PASSWORD=${ENA_POSTGRES_PASSWORD}
- ENA_USERNAME=${ENA_USERNAME}
- ENA_PASSWORD=${ENA_PASSWORD}
- ENA_USE_DEV_ENDPOINT=${ENA_USE_DEV_ENDPOINT}
- ENA_UPLOAD_FREQ_SECS=${ENA_UPLOAD_FREQ_SECS}
- ENA_TOKEN=${ENA_TOKEN}
depends_on:
- db
db:
image: postgres:15-bookworm
hostname: db
restart: unless-stopped
env_file: .env
volumes:
- pg_data:/var/lib/postgresql/data
- ./scripts/init_ena_db.sh:/docker-entrypoint-initdb.d/init_ena_db.shIn the .env you need to configure the following values:
ENA_POSTGRES_DB=ena
ENA_POSTGRES_USER=ena
ENA_POSTGRES_PASSWORD=changeme
ENA_USERNAME=Webin-xxxxx
ENA_PASSWORD=xxxxxxxxxxx
ENA_USE_DEV_ENDPOINT=True
ENA_UPLOAD_FREQ_SECS=5
ENA_TOKEN=changemeThe database need to initialize the ena database with the correct username and password. This can be achieved using the following script as the /docker-entrypoint-initdb.d/init_ena_db.sh:
#!/bin/bash
set -e
psql -v ON_ERROR_STOP=1 --username "$POSTGRES_USER" --dbname "$POSTGRES_DB" <<-EOSQL
CREATE USER "$ENA_POSTGRES_USER";
ALTER USER "$ENA_POSTGRES_USER" PASSWORD '$ENA_POSTGRES_PASSWORD';
CREATE DATABASE "$ENA_POSTGRES_DB";
GRANT ALL PRIVILEGES ON DATABASE "$ENA_POSTGRES_DB" TO "$ENA_POSTGRES_USER";
EOSQLcurl 'https://domain.com/api/jobs/study/' \
-H 'Authorization: token xxxxxxxxxxxxxx' \
-H 'Content-Type: application/json' \
-d '{
"template": "default",
"data": {}
}' \
curl 'http://domain.com/api/jobs/ser/' \
-H 'Authorization: token xxxxxxxxxxxxxx' \
-H 'Content-Type: application/json' \
-d '{
"template": "default",
"data": {"sample": {"host subject id": "xy1234"}},
"files": ["/data/example.cram"]
}'curl 'http://domain.com/api/analysisjobs/' \
-H 'Authorization: token xxxxxxxxxxxxxx' \
-H 'Content-Type: application/json' \
-d '{
"job": <job_id>,
"template": "default",
"data": {}
}'curl 'http://domain.com/api/analysisfiles/' \
-H 'Authorization: token xxxxxxxxxxxxxx' \
-H 'Content-Type: application/json' \
-d '{
"job": <analysis_job_id>,
"file_name": "/data/example.fa.gz",
"file_type": FASTA
}'curl 'http://domain.com/api/analysisjobs/<job_id>/enqueue' \
-H 'Authorization: token xxxxxxxxxxxxxx'curl http://domain.com/api/jobs/<job_id>/release/ \
-H 'Authorization: token xxxxxxxxxxxxxx'curl http://domain.com/api/analysisjobs/<job_id>/release/ \
-H 'Authorization: token xxxxxxxxxxxxxx'